Record Data
1) Record data description
In order to train SVM model, all the data except the label should be quantitative. The dataset used here is a subset of gathered data from clinicaltrials.gov API, which have four quantitative columns, AdverseEffectsorDeath, DurationDays, EnrollmentCount and ArmNumber, as well as one label column OverallStatus.
Record dataset
2) Models & Results for SVM with record data
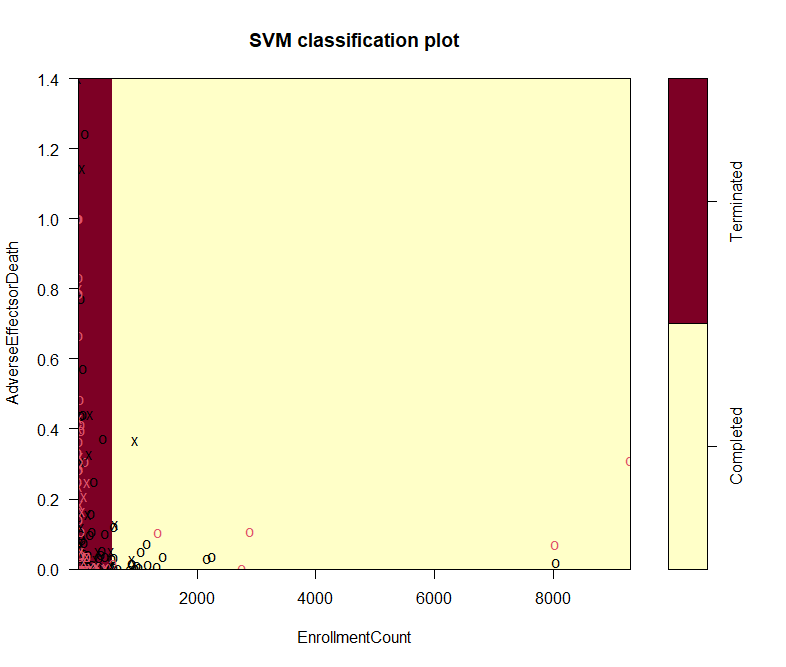
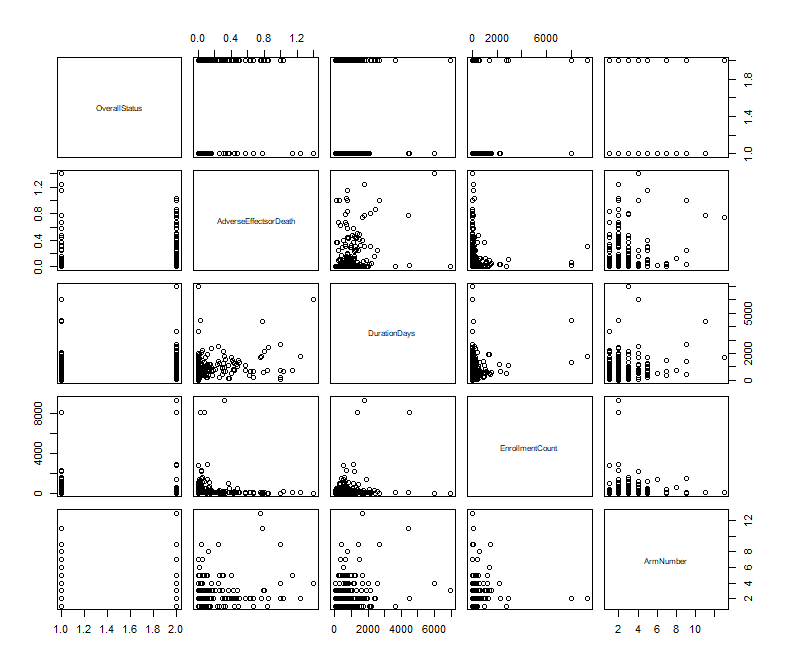
Figure 1 has shown a broad overview of correlation among all attributes in the dataset. It is hard to tell the general relationship among these graphs.
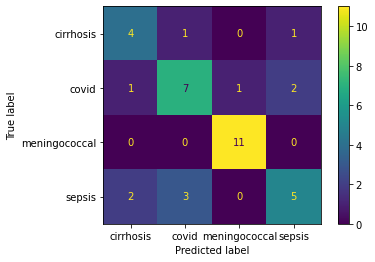
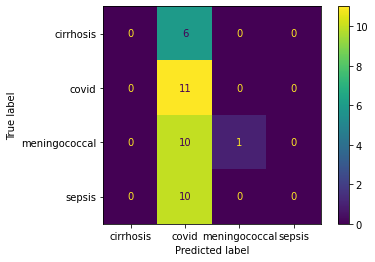
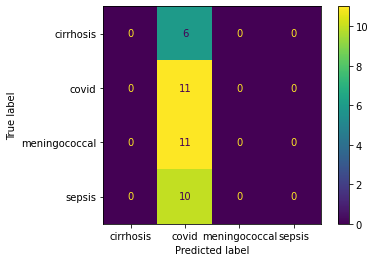
Train the data with polynomial SVM, it has the accuracy of 43%. The confusion matrix has suggest the low accuracy on predicting the completed trials, 13%, and a median accuracy on predicting the terminated trials, 72%. What is more, the models trained with linear SVM and radial SVM have similar accuracy at 48% and 43%, respectively.



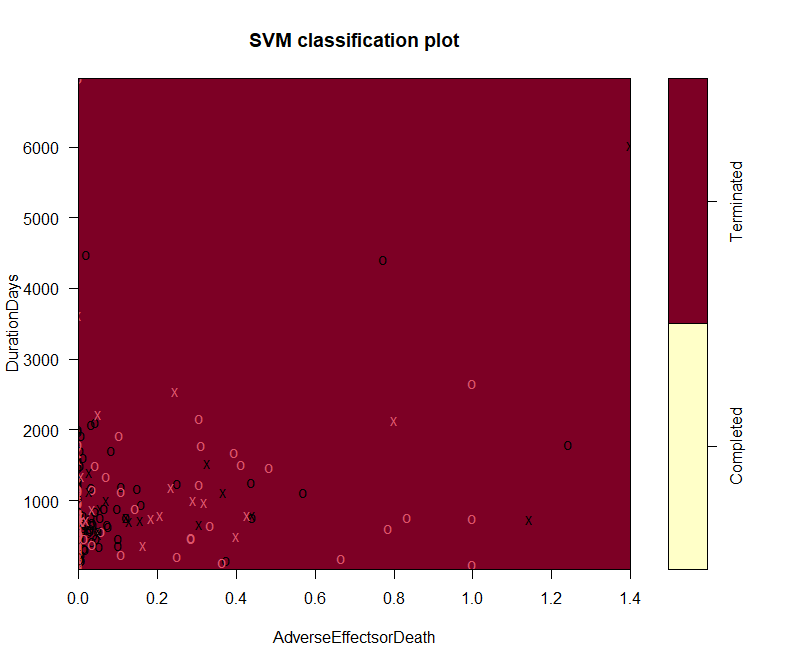
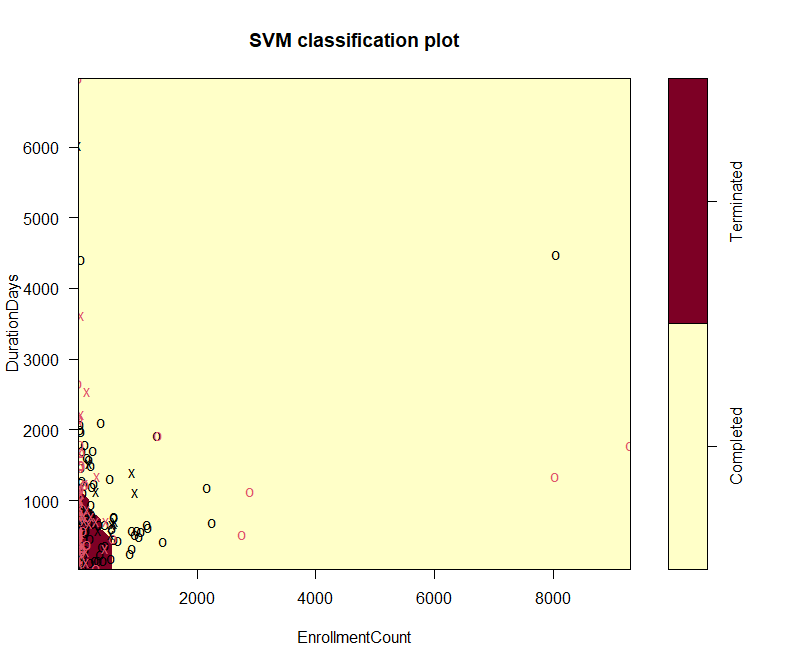
This part presents more detailed plots of four variables. Figure 5 presents the classification of labels along with the columns of AdverseEffectsorDeath vesus DurationDays. This plot does not show clear correlation between these two columns. A large proportion of data are lying within the classification of completed trials, though they should be around evenly distributed into both parts. Figure 6 shows the different distribution of labels, based on prediction, in the correlation plot of EnrollmentCount vesus DurationDays. Similarly, this graph does not suggest the these two factors have correlation, either.